



Playing with LEGO® made of DNA
(Italian version - Android app version)
(an educational webpage that demonstrates some concepts of DNA-based nanotechnology ... and lets you play around a bit )
copyright: Giampaolo Zuccheri 2021
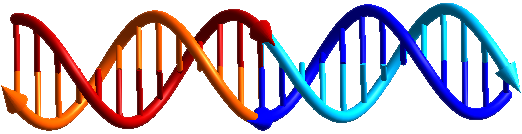
DNA is the molecule that codes life and is the same in all organisms. The chemistry and structure of DNA are perfect for encoding information that can lead to the specific bond between two molecules. Life itself depends on the effectiveness and specificity of these bonds.
Each DNA molecule is characterized by its sequence: it is like a word that describes its chemical structure. The sequences of two DNA molecules (or two sections of molecules) are said to be complementary when the information code they carry can mediate their specific interaction (i.e., it allows them to bind to each other). The DNA complementarity code was discovered by James Watson and Francis Crick. By exploiting the knowledge of this information code, as if they were a set of grammar rules, we can design the sequence of synthetic DNA molecules so that they can bind to each other. These molecules are made in the laboratory and do not necessarily have any biological function, but they can allow you to build "bricks." Then you can assemble different bricks together to make increasingly complex constructions, similarly to what we can do with LEGO® blocks. The difference between DNA-based nanotechnology and "normal" LEGO® is that the bricks and resulting constructions are extremely small (on the nanometer scale) and that no hands are needed to build the bricks or to assemble the constructions. Everything happens automatically: you just need to design the right sequences, synthesize them and dissolve them all together in an aqueous solution. Then they will specifically bind to each other.
What are DNA LEGOs® for? For now there are not too many practical applications, but there are demonstrations that with the control of their assembly, their structure and their behavior it is possible to build innovative materials, produce more efficient catalysts (useful for polluting less) or even innovative drugs.
.
 |
A DNA molecule is a flexible chain, comparable to cooked spaghetti (but much thinner). In a solution, the molecules are very numerous and swim in random directions, sometimes meeting each other. If two DNA molecules with random sequence meet, they normally do not interact, as shown in the simulation below (the different colors represent different sequences). |  |
(press the green flag to restart the simulations or the STOP sign to stop it).
 |
But if instead the two molecules have complementary sequences and by chance they meet, they bind in a specific way. Furthermore, by binding, the two flexible "cooked spaghetti" molecules form a more rigid double helix (perhaps you have also seen it represented in school books or on other websites). In our graphic representations, dark sequences are complementary to light ones of the same color. See in the simulation below what happens when two complementary DNA molecules accidentally meet. In a solution, many DNA molecules with different sequences can be present: only the complementary ones bind when they meet, the others do not interact. |
(press the green flag to restart the simulations or the STOP sign to stop it).
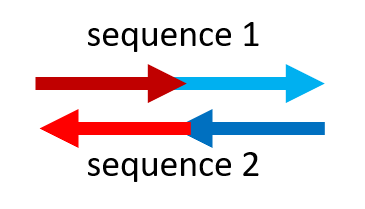
For the purposes of this demonstration, we can identify a stretch of DNA sequence (a 'word') with a letter, such as "A" or "a". In this symbology, we consider "A" complementary to "a" , "B" complementary to "b", and so on (uppercase letter complementary to the same lowercase letter). A DNA molecule (a piece of 'spaghetti') can have a sequence consisting of a single section or of several sections in a row (which we could represent, for example, with "AcB" showing that they are different sections). DNA molecules have chemical characteristics that cause them to bind to each other in an 'antiparallel' fashion, as if the head of a sequence is bound to the foot of its complementary sequence, and vice versa. Graphically, the sequences are represented as arrows with a point: in the double helixes formed by two complementary sequences, the points of the two arrows are oriented in the opposite way (see the drawing on the right). The 'sticks' that connect two DNA molecules are the chemical bonds that are established between two complementary molecules to hold them together in a helix. |
 |
If a molecule is made up of multiple sections of sequences one after the other, antiparallelism will have to be taken into account to design its complementary sequence (and so build the your first LEGO® brick). For example, "AB" will bind (is complementary) to "ba" and not to "ab" or "AB".
Now design your first brick : you have to get 2 complementary sequences: choose from the options with the selectors. Then, check if your answer is correct when the molecules meet in the simulation. The structure you want to obtain is this:
and we can schematize it as of below:
(press the green flag to restart the simulations or the STOP sign to stop it. Reload the web page to reset any of your choices).
To make LEGO® with DNA, branched structures are often used: such as 3- or 4-arm junctions. Using these, many different shapes can be obtained. These bricks are made by designing sequences of 3 or 4 different strands, each consisting of 2 sections, 2 by 2 complementary (as you did before).
Try designing sequences now (choosing your options with the selectors) to build this 3-arm junction:

schematized as:

(press the green flag to restart the simulations or the STOP sign to stop it. Reload the web page to reset any of your choices).
Now you know how to build some types of DNA bricks (actuially, there are many different ones). The fun in LEGO® is putting the blocks together to build something bigger. It is also done with DNA bricks! DNA bricks must be equipped with "cohesive ends" that bind one brick to another in a specific and automatic way.
Cohesive ends are short stretches of complementary sequences: when they meet they make double helixes bridging between two bricks and thus the bricks automatically assemble.
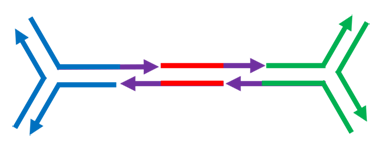
Now, we invite you to design the ends (the purple segents in the scheme) for assembling 3 DNA bricks. The goal is obtaining the structure shown below. Choose the right ends with the selectors.

schematized as:
(press the green flag to restart the simulations or the STOP sign to stop it. Reload the web page to reset any of your choices).
Congratulations! you have completed this demonstration on DNA-based nanotechnology.
If you have comments or are interested in learning more about the subject, write to Giampaolo Zuccheri.
You could also take a look at the video below (subtitles are available)
